Announcements
This section describes the format of and some features associated with NIRI data. More detailed information on reduction of NIRI data accompanies the NIRI IRAF package.
- undistort.pro - NIRI with Altair has been measured to have radial barrel distortion at f/32. More details here. The Undistort.pro IDL program will correct for this. PIs interested in high-precision astrometry are encouraged to contact either the NIRI or Altair instrument scientists to discuss this issue.
- Gemini IRAF Package
- NOTE: To browse what Gemini Users have exchanged about tricks, issues and solutions regarding reduction of NIRI data, please go to the DR forum (tag=NIRI).
Data primer
This page describes NIRI data and its reduction:
Raw data and nprepare
Raw NIRI images are written as multi-extension FITS (MEF) files with a single unnamed extension [1] containing the image data. Most of the information is contained in the primary header unit (PHU, extension [0]). Additional important information is added to the PHU by the IRAF task NPREPARE, which is part of the current release of the Gemini IRAF package.
Raw files stored by the Data Handling System (DHS) are written with file names of the form N20080101S0001.fits. The date is the UT date at the beginning of the night. Because the DHS is writing data from multiple instruments, including the wavefront sensors, the NIRI file number sequence is frequently interrupted. Missing file numbers do not necessarily indicate missing NIRI data.
NIRI writes images taken as coadds as a summed, not averaged, image. This means that if one takes 10 coadds of 1 sec each, the final image will have a flux level in ADU equivalent to 10 sec. The task NPREPARE leaves the pixel values as written by NIRI but modifies the header keyword EXPTIME to be the total exposure (the original exposure time multiplied by the number of coadds). NPREPARE adds a keyword COADDEXP that contains the original exposure time for each individual frame. While this behavior may be confusing to some, it is consistent with one ADU in the output image representing a fixed number of detected electrons.
The NIRI array controller averages the data based on the number of digital averages or multiple non-destructive read pairs, so no modification is necessary for different LNRS or NDAVGS values. The flux value will correspond to the exposure time, and the noise will vary as a function of the number of reads.
NPREPARE can help you better understand the raw data by adding a few header keywords. These include GAIN, RDNOISE, SATURATI, NONLINEA, and BIAS which give estimates of the gain (e-/ADU), read noise (e-), saturation level (ADU), the level at which the data start to be non-linear (ADU), and the array bias voltage. These important header keywords are used by other scripts in the NIRI package. NPREPARE refers to a data file named nprepare.dat that contains vital information for adding these header keywords. The user must make sure that the version of NPREPARE and nprepare.dat is appropriate for the data being reduced. nprepare.dat is included with the current release of the NIRI package. nprepare.dat will be modified from time to time to reflect updates in the array parameters or instrument configuration.
Note that the slit centerings are slightly different than before, and we have distributed a modified version of nsappwave.dat to account for a small central wavelength shift.
NPREPARE can also add variance and data quality planes, as described in the help pages for NPREPARE.
Array characteristics
Raw NIRI images are dominated by the landscape of the pixel sensitivity variations. You will notice a number of common features:
- small-scale curved "stripes" running vertically. These result from the manufacture of the array and flatten very nicely.
- a circular pattern of ripples resembling a thumbprint in the upper left quadrant. This will also flatten out.
- three small regions of bad pixels, about 10 pixels across, two of which have a partially bad column below them. These are left over from the procedure that excised "photon-emitting defects" from the array, and are totally dead. Dither patterns should be large enough to guarantee that these pixels can be corrected.
- a few hundred hot pixels, mostly in the upper left corner and in a clump near the bottom center
- a prominent crack in the array substrate, seen as a line of bad pixels in the lower right-hand corner
- an overall large-scale bright region in the center of the array with darker regions top and bottom
- offsets in overall sensitivity at the quadrant boundaries
In addition, the array has a frame that can be seen along the top and right edges of the frame, with rounded corners.
CLEANIR.py
The GNAAC detector controller in NIRI and GNIRS sometimes superimposes vertical striping, horizontal banding, and quadrant offsets on the data. The "cleanir" python routine can remove many of these artifacts. Run the script with no arguments to see the full help.
In the default mode cleanir assumes that the pattern noise in a quadrant can be represented by a fixed pattern which is repeated over the entire quadrant. The default size for this pattern is 16 pixels wide and 4 pixels high (which may be changed via the -x and -y flags). The pattern is determined for each quadrant by taking the median of the pixel value distribution at each position in the pattern (which may be visualized with the -g flag). Once the median pattern has been determined for a quadrant it is replicated to cover the entire quadrant and subtracted, and the mean of the pattern is added back to preserve flux. The standard deviation of all the pixels in the quadrant is compared to that before the pattern subtraction, and if no reduction was achieved the subtraction is undone. The pattern subtraction may be forced via the -f flag, which may be necessary if the pattern is low-amplitude. This process is repeated for all four quadrants and the cleaned frame is written to c
Cleanir can also clean each row independently (via the -r flag), which can be useful for removing row structures which are sometimes seen in GNIRS data. In this mode cleanir looks for 8-pixel periodicities in each row, subtracting off the median 8-pixel pattern and adding a constant to bring the median of the row up to the median level of the quadrant. This mode should be used with caution as it may be affected by features which cover a significant fraction of a row, e.g. extended objects, sky lines in GNIRS, or residual image from the GNIRS acquisition keyhole. The -r option is not recommended for GNIRS XD flats.
The GNAAC pattern noise is often accompanied by an offset in the bias values between the four quadrants. One may want to use the cleanir -q flag to try to remove this offset. This attempts to match the iteratively determined median value of each quadrant. This method works best with sky subtraction (i.e. with the -s flag), and does not work well if there are large extended objects in the frame. By default the median is determined from the entire frame, although one can use the -c flag to only use a central portion of the image. Note that the derived quadrant offsets will be applied to the output pattern file.
Cleanir understands glob expansions, however, the entire string must be quoted or any pattern matching characters (*,?) must be escaped with a backslash to avoid being interpreted by the shell.
Options
-a : use all pixels for pattern determination
-b <badpixelmask> : specify a bad pixel mask (overrides DQ plane)
-c <frac> : use central <fraction> of image for bias adjustment [1]
-d <dir> : specify an input data directory
-f : force cleaning of all quads even if stddev does not decrease
-g # : graph results (0=none, 1=pattern, 2=offsets, 3=both)
-h <val> : ignore values above <val> in pattern determination
-m : use median instead of fitting a Gaussian
-o <file> : write output to <file> (instead of c<infile>)
-p <file> : write full-frame pattern to <file>
-q : adjust quadrant offsets
-r : row filtering (useful for GNIRS XD spectra)
-s <sky> : sky frame to help in pattern recognition
-v : verbose debugging output
-x <size> : set pattern x size in pix [16]
-y <size> : set pattern y size in pix [4]
Version
2019-May-07
GNIRS
Cleanir appears to work well on raw GNIRS XD files and sky-subtracted XD and long slit files. It does not work as well on raw long slit files. You may need to try several options to get the best cleaning of your data:
1. cleanir -q filename
2. cleanir -fq filename
3. cleanir -rq filename
The first example should always be attempted first as it is the most robust and least likely to introduce additional artifacts into your data. The second example uses the -f flag to force the pattern subtraction. Example 3 uses the -r flag for row-filtering, and should be the last resort if the first two methods are not successful at removing the noise.
To clean sky-subtracted GNIRS frames a sky-subtracted image should first be produced using gemarith to preserve the MEF structure:
gemarith filename1 - filename2 filename3
cleanir -q filename3
NIRI
Removing the pattern from NIRI spectroscopy can also be difficult because of many vertical sky lines. By default f/6 spectroscopy with the 2-pixel or blue slits (which do not fill the detector), uses the empty regions at the bottom (1-272) and top (720-1024) of the array for measuring the pattern. This is not possible for other modes of spectroscopy where the spectrum fills the detector. For these modes it is best to do sky subtraction before pattern removal. The quickest method is to pass a sky frame (or an offset frame) via the -s flag. The manual method is to generate and subtract the sky, determine and save the pattern via the -p flag, then subtract the pattern from the original image. One may use the -a flag to force using all of the pixels for the pattern determination.
Examples
- Remove pattern noise from an image using the default parameters:
INPUT cleanir.py N20100101S0001.fits
OUTPUT: cN20100101S0001.fits - Remove pattern noise, saving the clean image as "clean.fits" and the pattern as "pattern.fits":
INPUT: cleanir.py -o clean.fits -p pattern.fits N20100101S0001.fits - Remove pattern noise from all quadrants even if the standard deviation does not improve:
INPUT: cleanir.py -f N20100101S0001.fits - Remove pattern noise and adjust the quadrant offsets to a common value:
INPUT: cleanir.py -q N20100101S0001.fits - Remove pattern noise using an offset frame as a sky to help with pattern determination:
INPUT: cleanir.py -s N20100101S0002.fits N20100101S0001.fits - Remove pattern noise using a bad-pixel mask to mask out extended objects:
INPUT: cleanir.py -b N20100101S0001msk.pl N20100101S0001.fits - Remove pattern noise from GNIRS XD spectra using row filtering and fixing quadrant offsets:
INPUT: cleanir.py -rq N20100101S0001 - Clean all frames in the "data" directory:
INPUT: cleanir.py "data/*.fits"
Requirements:
Download:
Cleanir is under development. Make sure you use the latest version.
Comments and suggestions are welcome and may be sent to astephens at gemini dot edu.
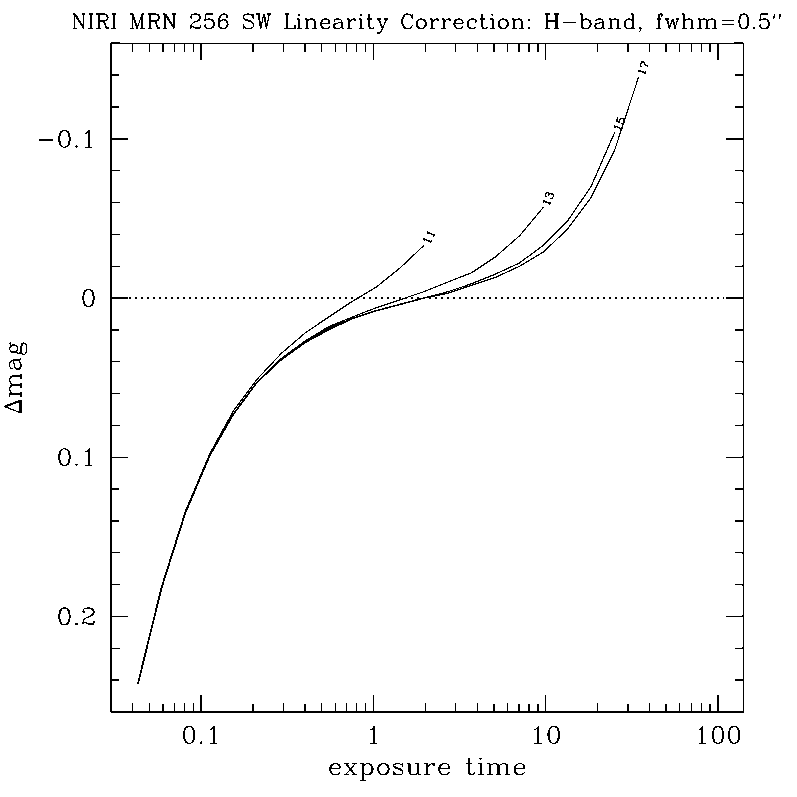
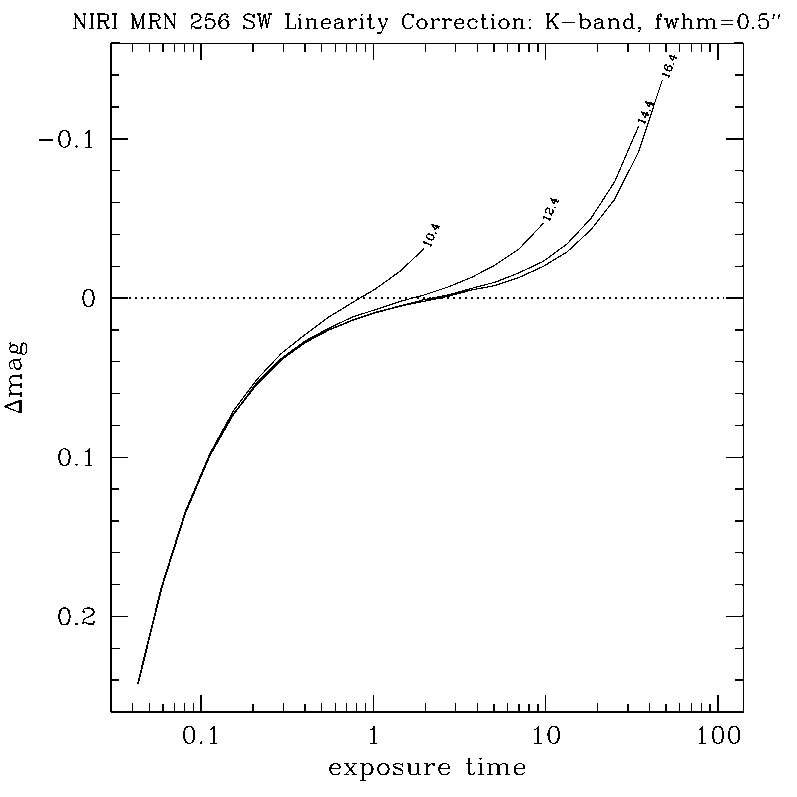
NIRLIN - NIR LINearization
NIRLIN should be used to linearize all science data. This version uses three coefficients to correct for non-linearity in the NIRI detector: an exposure time correction, a counts squared term, and a counts cubed term. These coefficients are dependent on the read mode and detector ROI. We have currently derived coefficients for the following configurations:
| Read Mode | ROI | Well Depth | dt | c2 | c3 |
| Low RN | 1024 | shallow | 1.266 | 7.39e-06 | 1.94e-10 |
| Medium RN | 1024 | shallow | 0.094 | 3.43e-06 | 4.81e-10 |
| Medium RN | 256 | shallow | 0.0103 | 6.82e-06 | 2.13e-10 |
| High RN | 1024 | shallow | 0.0097 | 3.04e-06 | 4.64e-10 |
| High RN | 1024 | deep | 0.0077 | 3.58e-06 | 1.82e-10 |
Note: We have not yet quantified the effect of linearizing flat fields.
USAGE:
NAME: nirlin.py - NIR linearization
SYNOPSIS: nirlin.py [options] infile
DESCRIPTION: Run on raw or nprepared Gemini NIRI data. This script calculates and applies a per-pixel linearity correction based on the counts in the pixel, the exposure time, the read mode, the bias level, and the ROI. Pixels over the maximum correctable value are set to BADVAL unless given the force flag. Note that you may use the glob expansion in the infile; however, any pattern matching characters (*,?) must either be quoted or excaped with a backslash.
OPTIONS:
-b : value to assign to uncorrectable pixels [0]
-f : force correction on all pixels
-0 <file> : write output to <file> [l<inputfile>]. If no .fits is included this is assumed to be a directory.
-v : verbose debugging output
VERSION: 2019 Aug 29
REQUIREMENTS:
Download the latest version of nirlin.py here.
Download the associated README file here.
DETAILS:
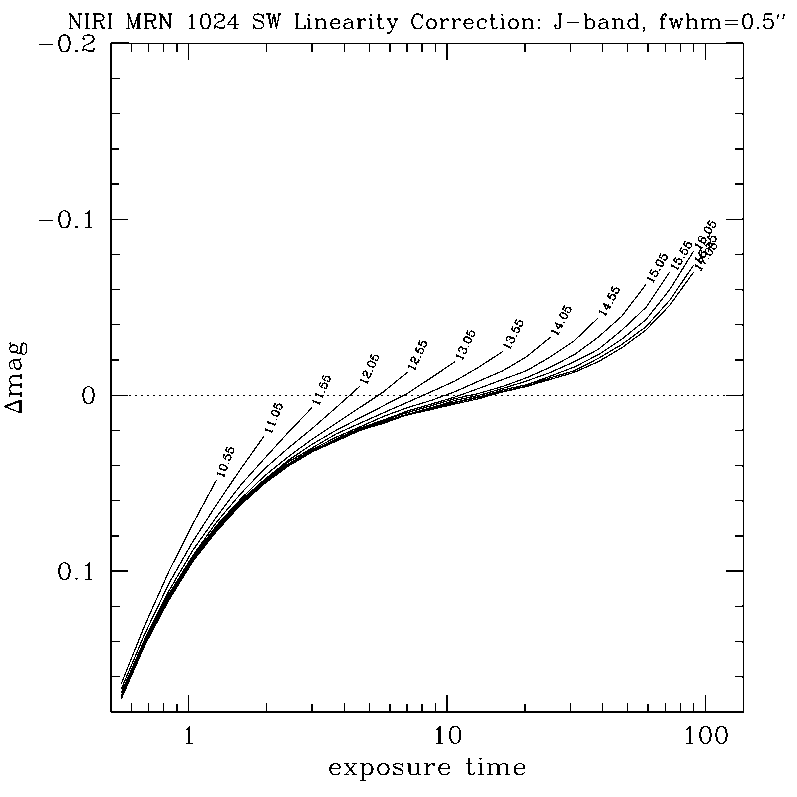
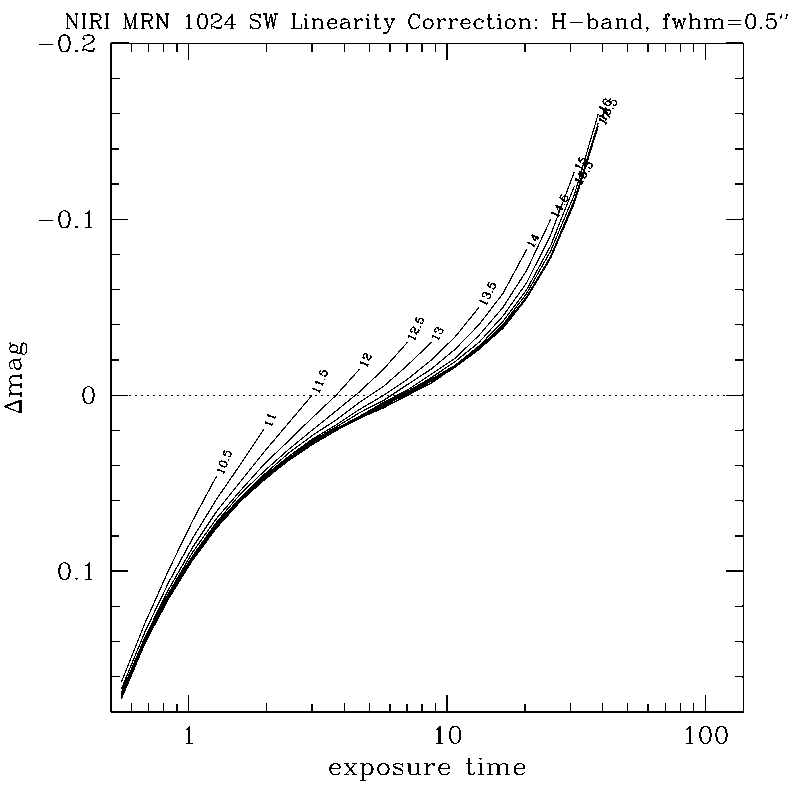
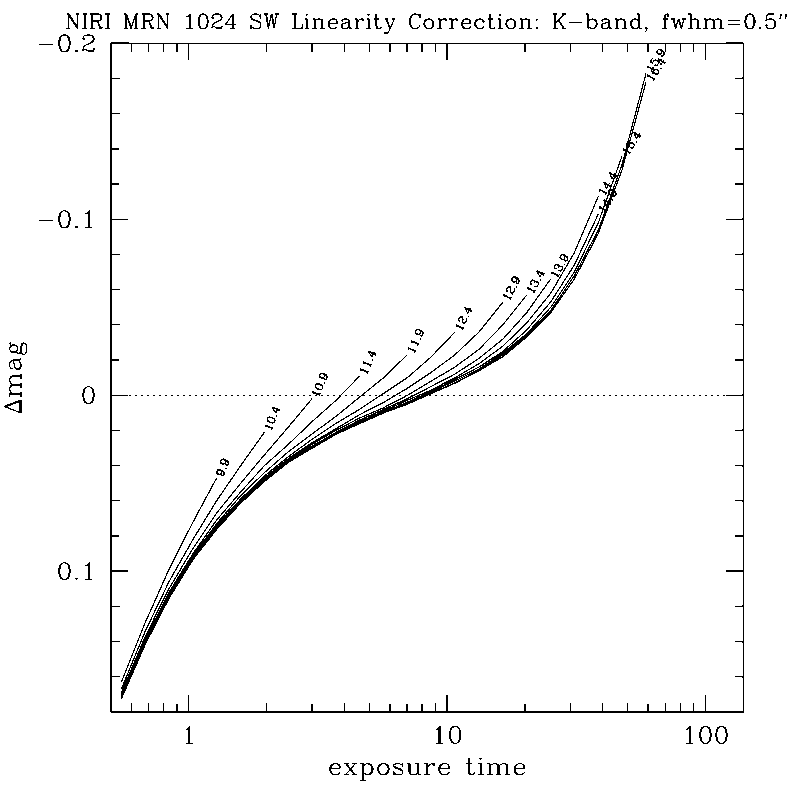
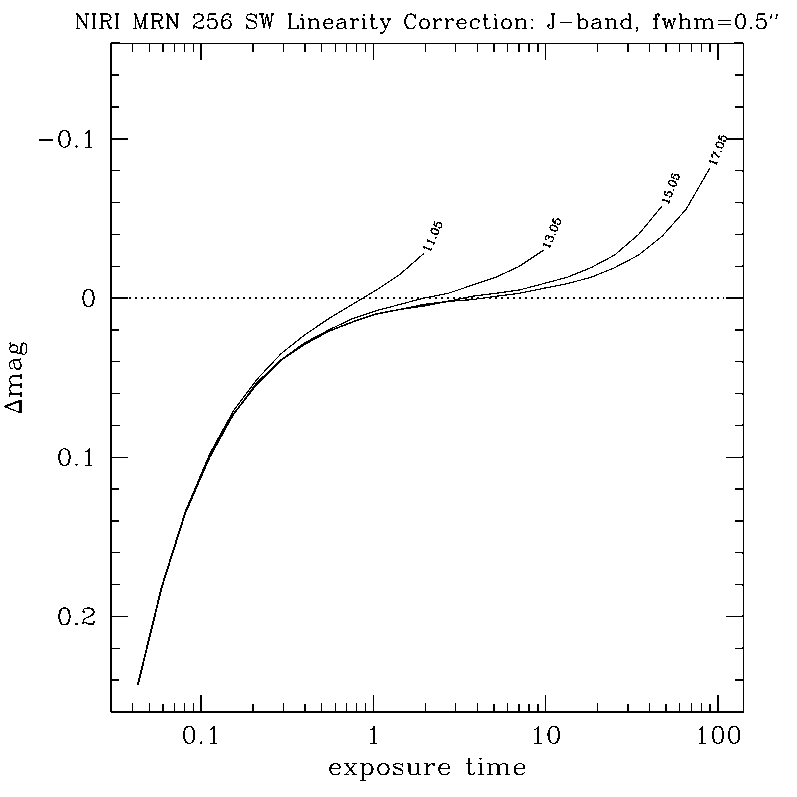
Any questions, comments or suggestions for improvements are welcome, and should be sent to Andrew Stephens (astephens at gemini dot edu).